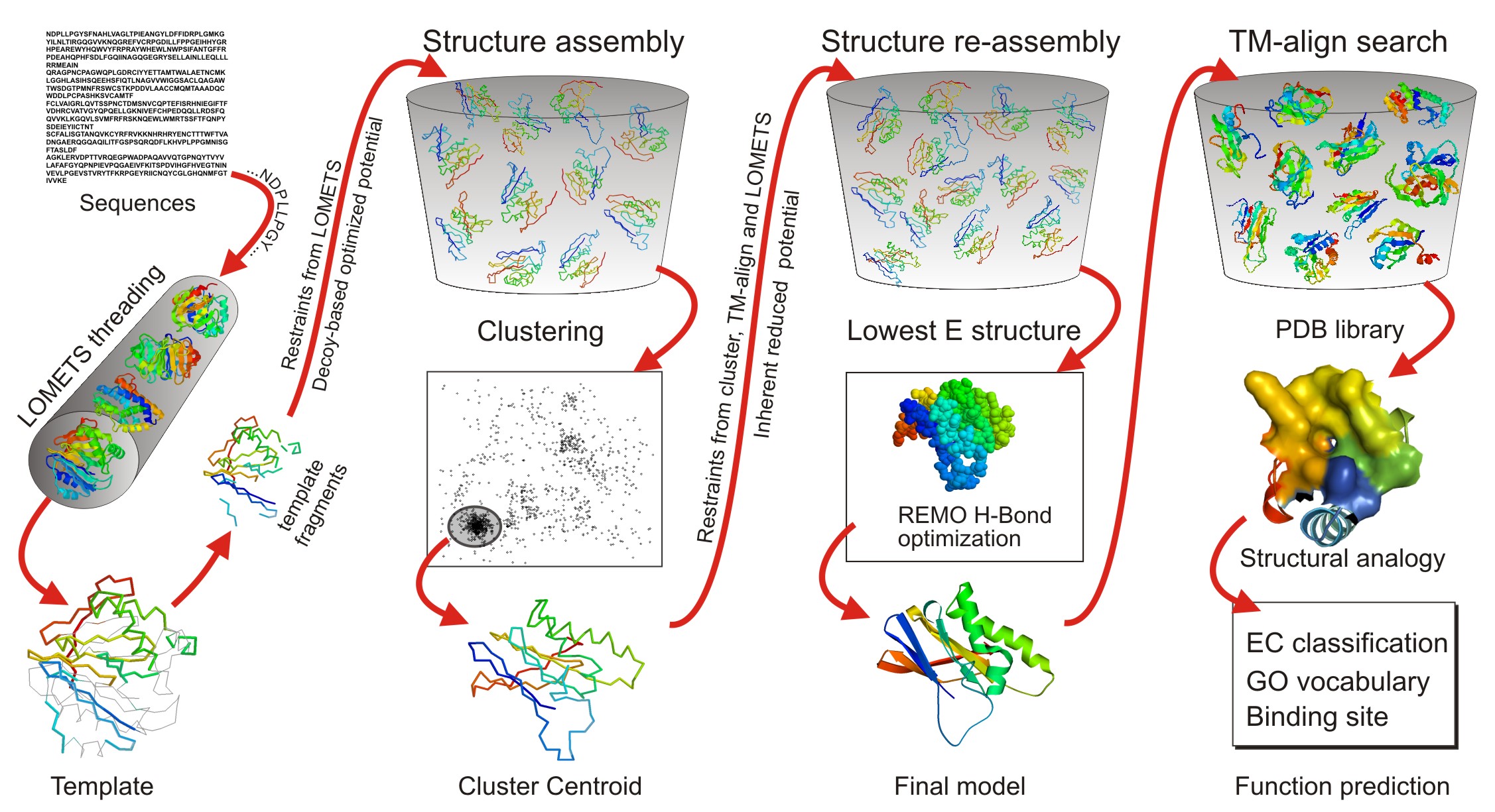
Integration of QUARK and I‐TASSER for Ab Initio Protein Structure Prediction in CASP11 - Zhang - 2016 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
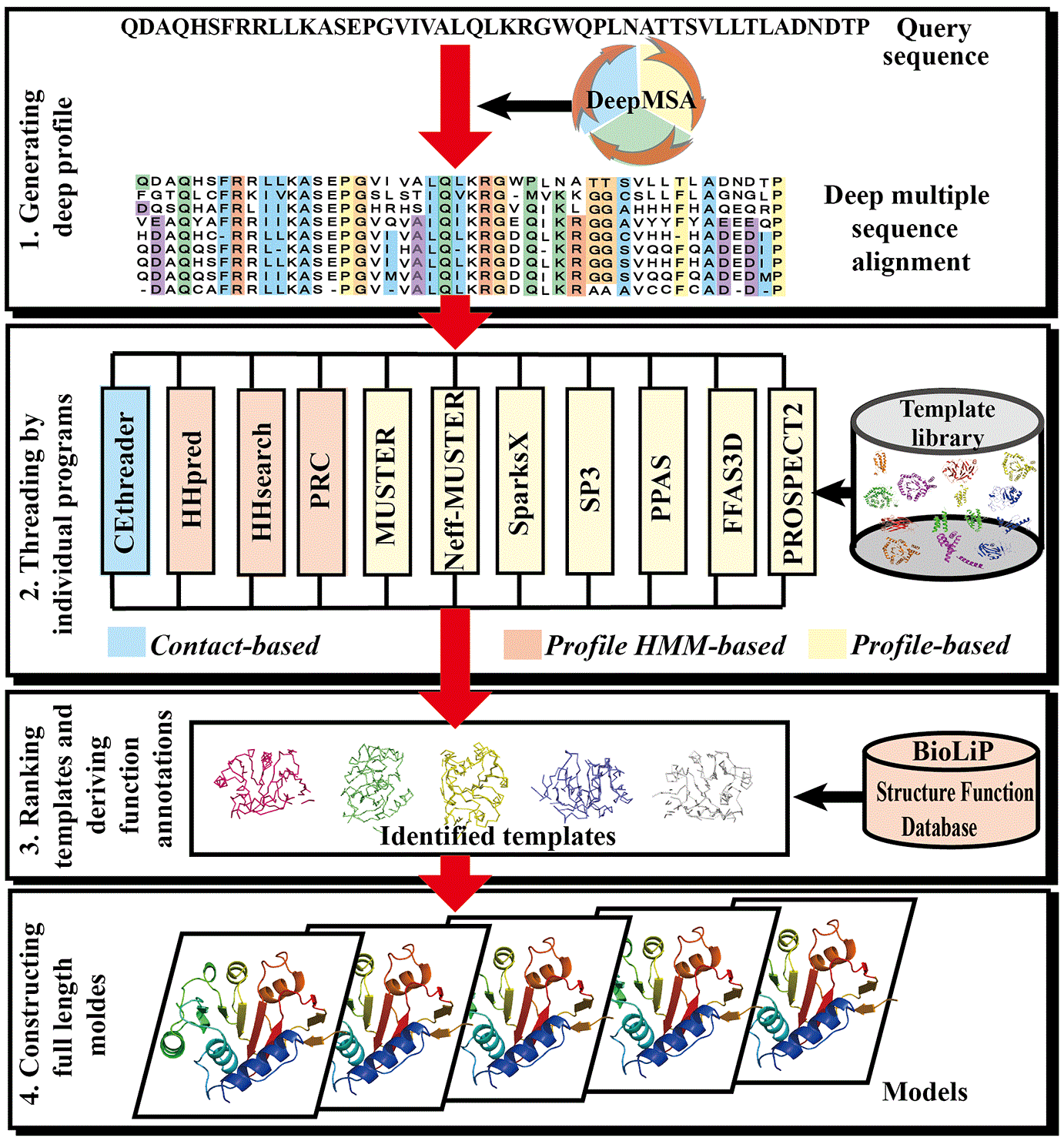
![PDF] I-TASSER server: new development for protein structure and function predictions | Semantic Scholar PDF] I-TASSER server: new development for protein structure and function predictions | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/682fc6a2dcf6edaadacb1a2f90fdd137aef28d15/2-Figure1-1.png)
PDF] I-TASSER server: new development for protein structure and function predictions | Semantic Scholar

Overview of the RBO Aleph structure prediction pipeline: the server... | Download Scientific Diagram

Interplay of I‐TASSER and QUARK for template‐based and ab initio protein structure prediction in CASP10 - Zhang - 2014 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
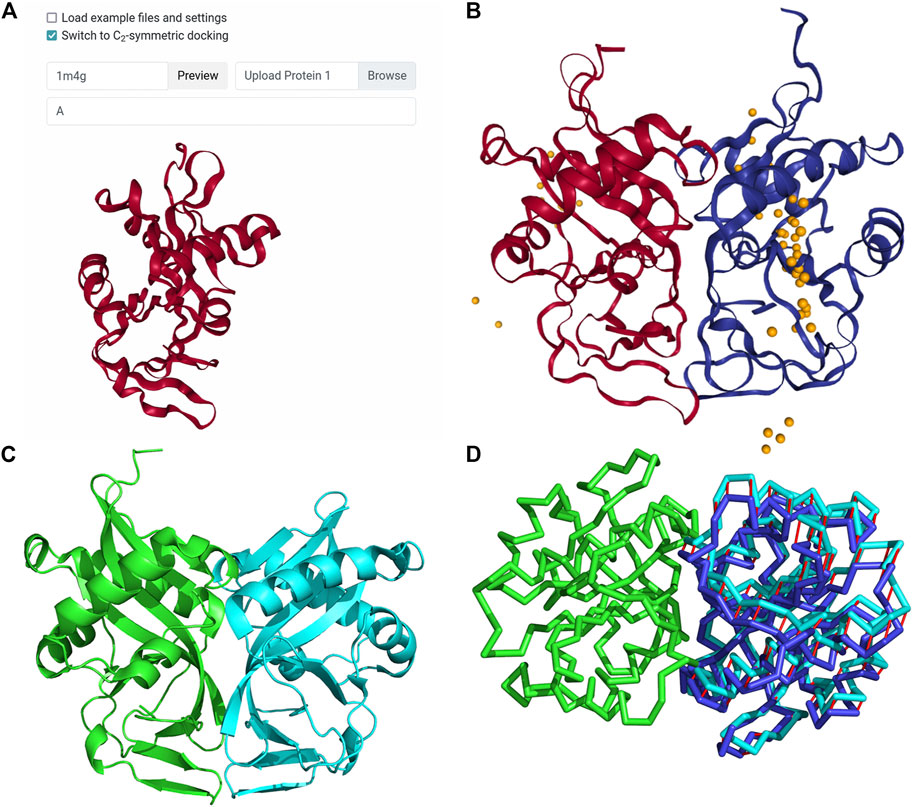
Frontiers | LZerD Protein-Protein Docking Webserver Enhanced With de novo Structure Prediction | Molecular Biosciences

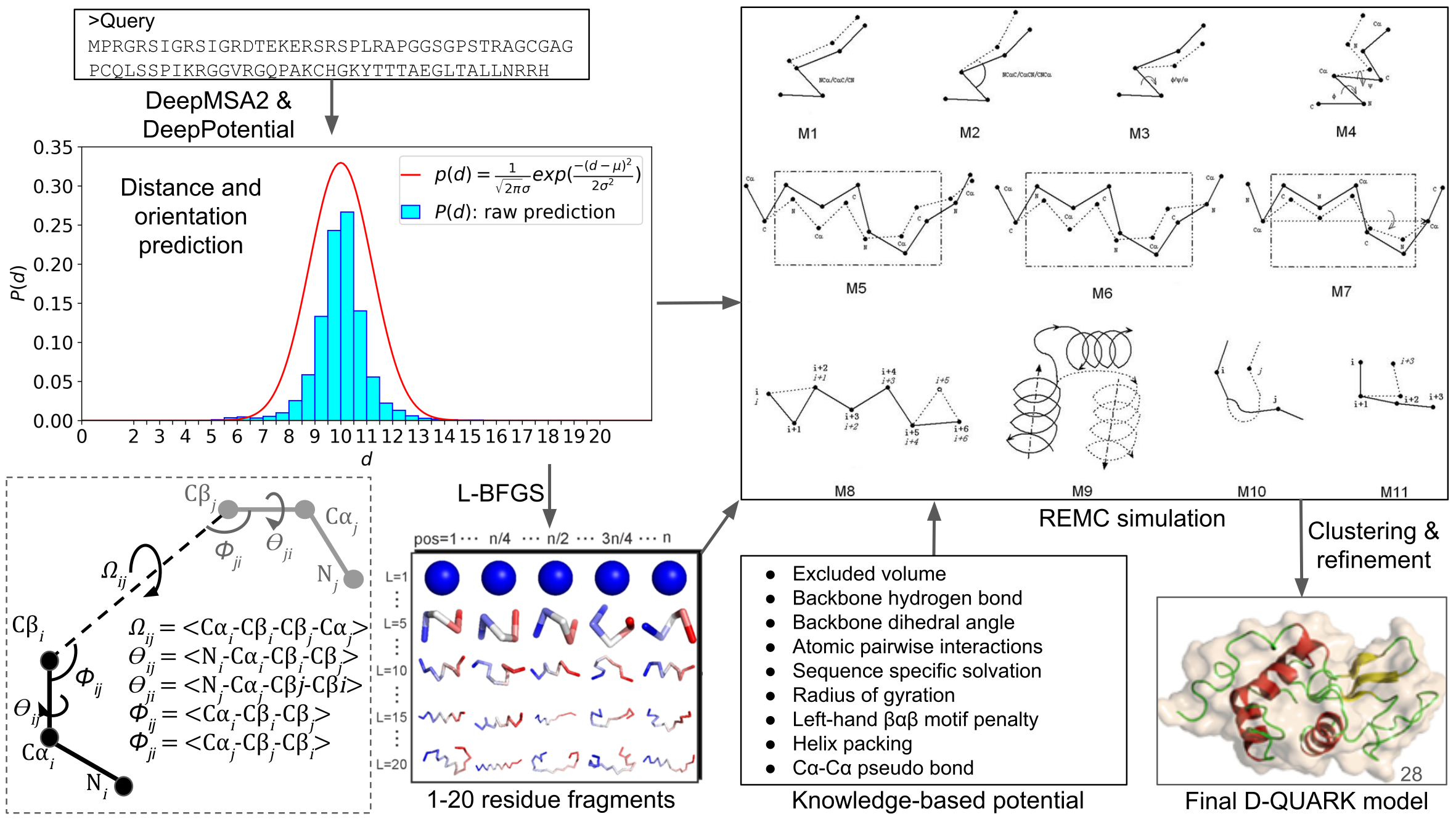
Examples of successful QUARK modeling results on known Hard E. coli... | Download Scientific Diagram
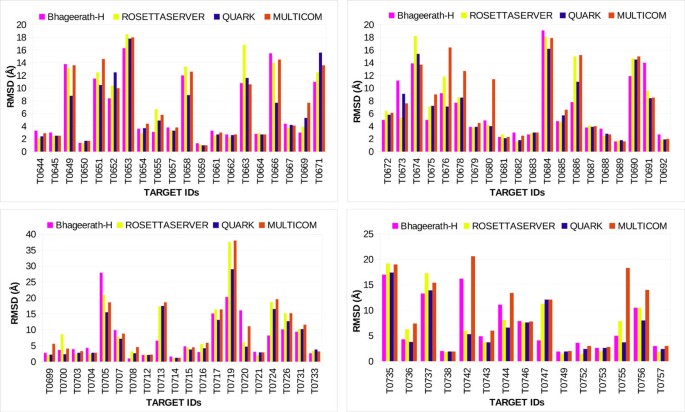
Bhageerath-H: A homology/ab initio hybrid server for predicting tertiary structures of monomeric soluble proteins | BMC Bioinformatics | Full Text

Use of the UNRES force field in template-assisted prediction of protein structures and the refinement of server models: Test with CASP12 targets - ScienceDirect

Computational generation of proteins with predetermined three-dimensional shapes using ProteinSolver - ScienceDirect
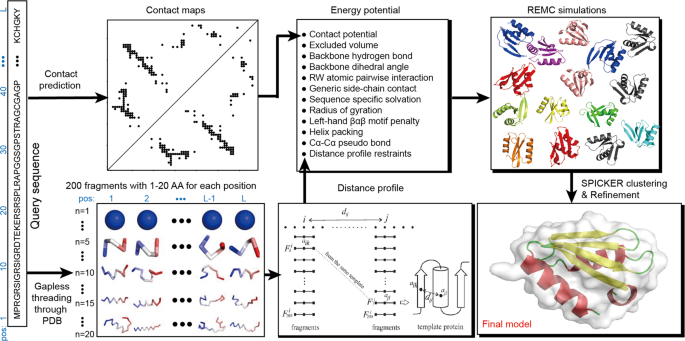
Improving fragment-based ab initio protein structure assembly using low-accuracy contact-map predictions | Nature Communications

Integration of QUARK and I‐TASSER for Ab Initio Protein Structure Prediction in CASP11 - Zhang - 2016 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

Deep‐learning contact‐map guided protein structure prediction in CASP13 - Zheng - 2019 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
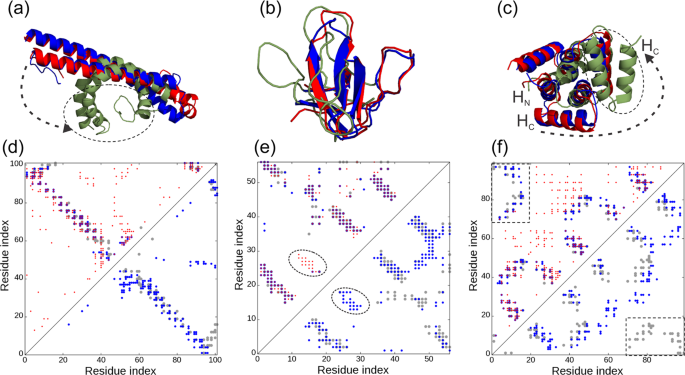
Improving fragment-based ab initio protein structure assembly using low-accuracy contact-map predictions | Nature Communications

Protein structure, amino acid composition and sequence determine proteome vulnerability to oxidation‐induced damage | The EMBO Journal